研究副教授 |生命科学学院, 系统生物学系
2024 南方科技大学,生命科学学院,系统生物学系,研究副教授
2020 南方科技大学,生物系/前沿与交叉科学研究院,研究副教授
2017 南方科技大学,生物系/前沿与交叉科学研究院,研究助理教授
2013 德国亥姆霍兹联合会马克斯-德尔布吕克分子医学中心,博士后
2013 柏林洪堡大学,细胞生物学博士
2007 浙江大学,药学院,药理学硕士
2005 浙江大学,药学学士&生物技术学士
个人简介
研究领域
RNA生物学、细胞生物学、肿瘤生物学
技术平台
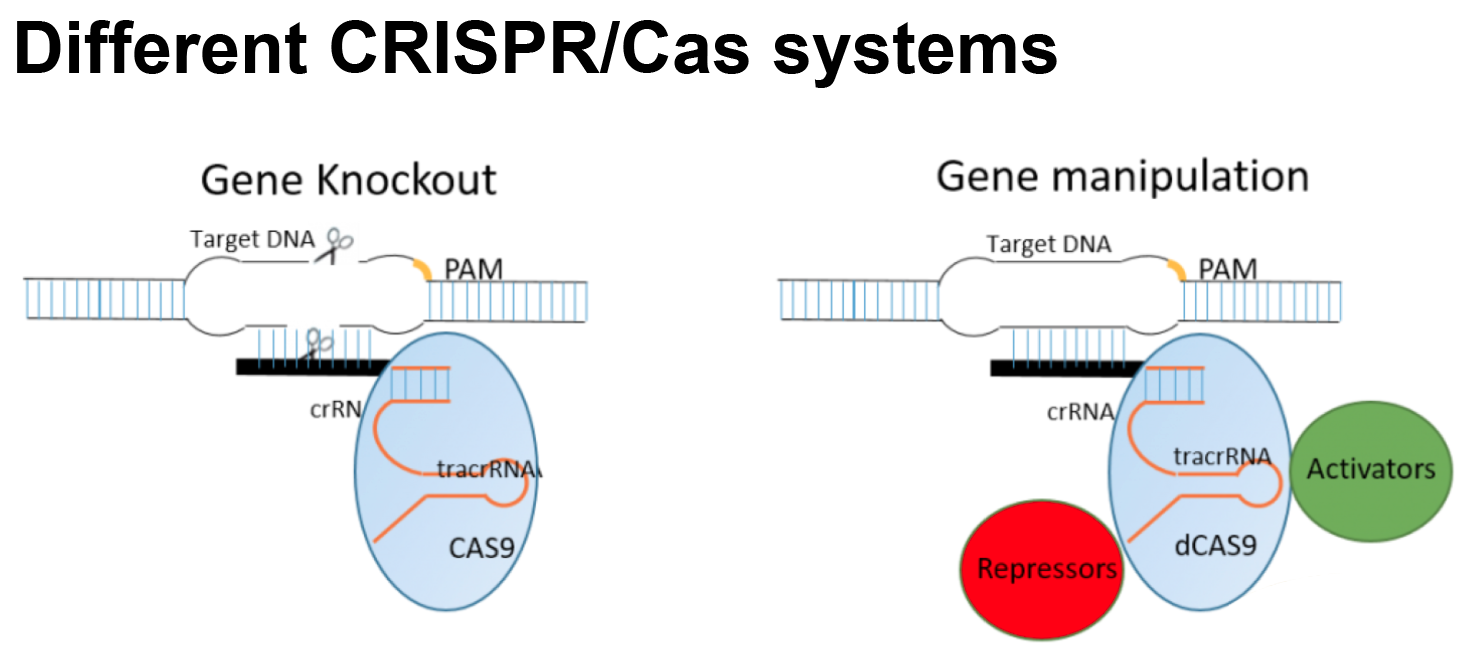
1. CRISPR基因筛选;
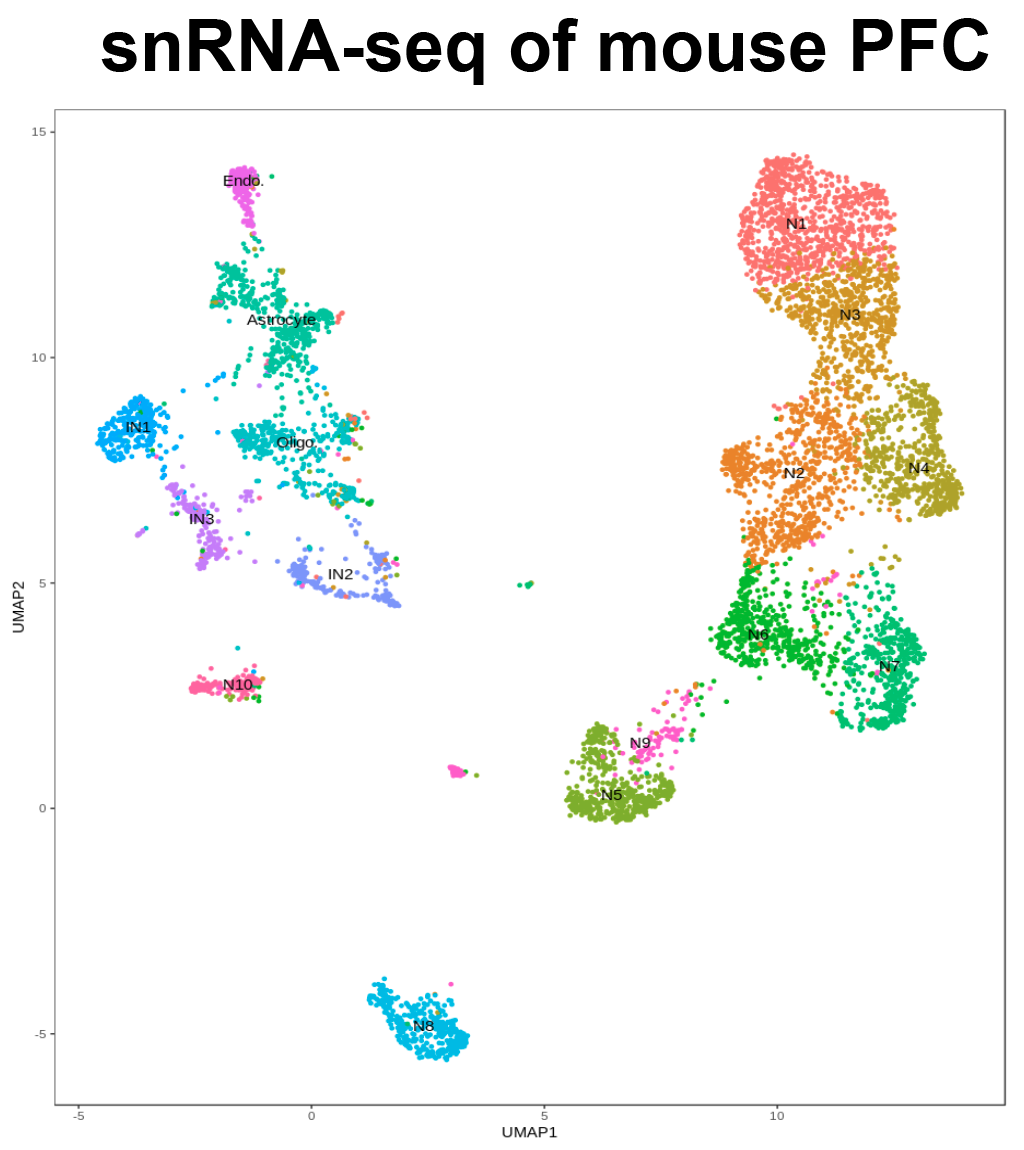
2. 单细胞测序;
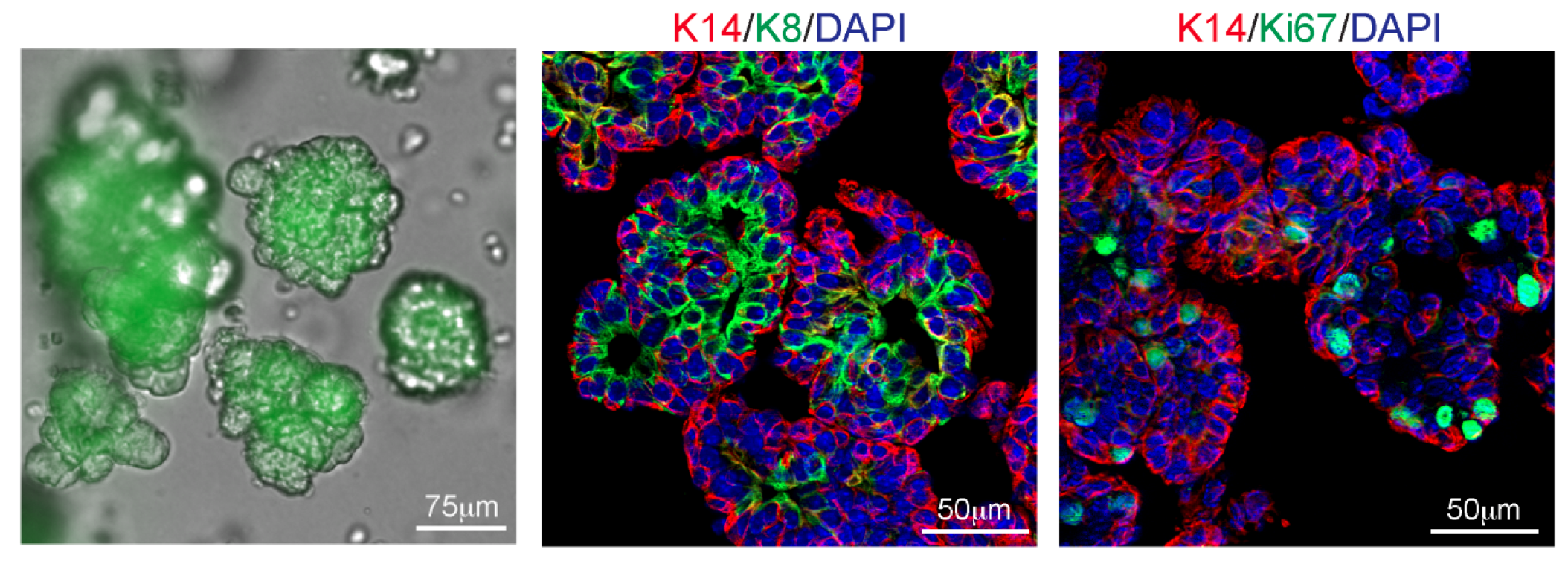
3. 类器官培养;
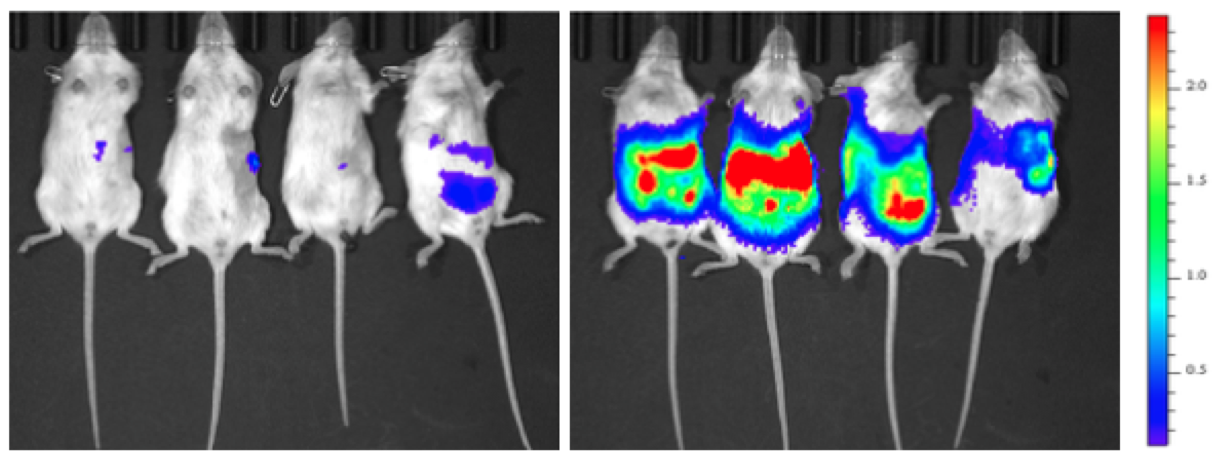
4. 转基因小鼠。
学术成果 查看更多
2024
Fang, L.*#, L. Zhang*, M. Wang*, Y. He, J. Yang, Z. Huang, Y. Tan, K. Fang, J. Li, Z. Sun, Y. Li, Y. Tang, W. Liang, H. Cui, Q. Zhu, Z. Wu, Y. Li, Y. Hu and W. Chen# (2024). “Pooled CRISPR Screening Identifies P-Bodies as Repressors of Cancer Epithelial-Mesenchymal Transition.” Cancer Res 84(5): 659-674.
2023
Zhu, Q.*#, X. Zhao*, Y. Zhang*, Y. Li*, S. Liu*, J. Han, Z. Sun, C. Wang, D. Deng, S. Wang, Y. Tang, Y. Huang, S. Jiang, C. Tian, X. Chen, Y. Yuan, Z. Li, T. Yang, T. Lai, Y. Liu, W. Yang, X. Zou, M. Zhang, H. Cui, C. Liu, X. Jin, Y. Hu, A. Chen, X. Xu, G. Li, Y. Hou, L. Liu#, S. Liu#, L. Fang#, W. Chen# and L. Wu# (2023). “Single cell multi-omics reveal intra-cell-line heterogeneity across human cancer cell lines.” Nat Commun 14(1): 8170.
Song, X., P. Guo, K. Xia, M. Wang, Y. Liu, L. Chen, J. Zhang, M. Xu, N. Liu, Z. Yue, X. Xu, Y. Gu, G. Li, M. Liu, L. Fang, X. W. Deng and B. Li (2023). “Spatial transcriptomics reveals light-induced chlorenchyma cells involved in promoting shoot regeneration in tomato callus.” Proc Natl Acad Sci U S A 120(38): e2310163120.
2022
Yang, C., Y. Liu, Y. Hu, L. Fang, Z. Huang, H. Cui, J. Xie, Y. Hong, W. Chen, N. Xiao, Q. Li, W.-H. Liu and C. Xiao (2022). “Myc inhibition tips the immune balance to promote antitumor immunity.” Cellular & Molecular Immunology.
Wang, W., H. Huang, H. Jiang, C. Tian, Y. Tang, D. Gan, X. Wen, Z. Song, Y. He, X. Ou and L. Fang (2022). “A Cross-Tissue Investigation of Molecular Targets and Physiological Functions of Nsun6 Using Knockout Mice.” Int J Mol Sci 23(12).
Cui, H., H. Yi, H. Bao, Y. Tan, C. Tian, X. Shi, D. Gan, B. Zhang, W. Liang, R. Chen, Q. Zhu, L. Fang, X. Gao, H. Huang, R. Tian, S. R. Sperling, Y. Hu and W. Chen (2022). “The SWI/SNF chromatin remodeling factor DPF3 regulates metastasis of ccRCC by modulating TGF-beta signaling.” Nat Commun 13(1): 4680.
Zou, X., B. Schaefke, Y. Li, F. Jia, W. Sun, G. Li, W. Liang, T. Reif, F. Heyd, Q. Gao, S. Tian, Y. Li, Y. Tang, L. Fang, Y. Hu and W. Chen (2022). “Mammalian splicing divergence is shaped by drift, buffering in trans, and a scaling law.” Life Sci Alliance 5(4).
Wang, D., Q. Mai, X. Yang, X. Chi, R. Li, J. Jiang, L. Luo, X. Fang, P. Yun, L. Liang, G. Yang, K. Song, L. Fang, Y. Chen, Y. Zhang, Y. He, N. Li and Y. Pan (2022). “Microduplication of 16p11.2 locus Potentiates Hypertrophic Obesity in Association with Imbalanced Triglyceride Metabolism in White Adipose Tissue.” Mol Nutr Food Res 66(5): e2100241.
Tian, S., B. Zhang, Y. He, Z. Sun, J. Li, Y. Li, H. Yi, Y. Zhao, X. Zou, Y. Li, H. Cui, L. Fang, X. Gao, Y. Hu and W. Chen (2022). “CRISPR-iPAS: a novel dCAS13-based method for alternative polyadenylation interference.” Nucleic Acids Res 50(5): e26.
Gao, X., J. You, Y. Gong, M. Yuan, H. Zhu, L. Fang, H. Zhu, M. Ying, Q. He, B. Yang and J. Cao (2022). “WSB1 regulates c-Myc expression through beta-catenin signaling and forms a feedforward circuit.” Acta Pharm Sin B 12(3): 1225-1239.
2021
Zhou, P., Y. Qi, X. Fang, M. Yang, S. Zheng, C. Liao, F. Qin, L. Liu, H. Li, Y. Li, E. Ravindran, C. Sun, X. Wei, W. Wang, L. Fang, D. Han, C. Peng, W. Chen, N. Li, A. M. Kaindl and H. Hu (2021). “Arhgef2 regulates neural differentiation in the cerebral cortex through mRNA m(6)A-methylation of Npdc1 and Cend1.” iScience 24(6): 102645.
Long, Y., Z. Liu, J. Jia, W. Mo, L. Fang, D. Lu, B. Liu, H. Zhang, W. Chen and J. Zhai (2021). “FlsnRNA-seq: protoplasting-free full-length single-nucleus RNA profiling in plants.” Genome Biol 22(1): 66.
Kamdem, N., Y. Roske, D. Kovalskyy, M. O. Platonov, O. Balinskyi, A. Kreuchwig, J. Saupe, L. Fang, A. Diehl, P. Schmieder, G. Krause, J. Rademann, U. Heinemann, W. Birchmeier and H. Oschkinat (2021). “Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling.” Magn. Reson. 2(1): 355-374.
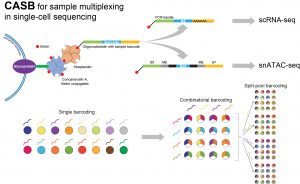
Fang, L.*#, G. Li#, Z. Sun, Q. Zhu, H. Cui, Y. Li, J. Zhang, W. Liang, W. Wei, Y. Hu and W. Chen* (2021). “CASB: a concanavalin A-based sample barcoding strategy for single-cell sequencing.” Mol Syst Biol 17(4): e10060.
Wu, X., Y. Zheng, M. Liu, Y. Li, S. Ma, W. Tang, W. Yan, M. Cao, W. Zheng, L. Jiang, J. Wu, F. Han, Z. Qin, L. Fang, W. Hu, Z. Chen* and X. Zhang* (2021). “BNIP3L/NIX degradation leads to mitophagy deficiency in ischemic brains.” Autophagy 17(8): 1934-1946.
2020
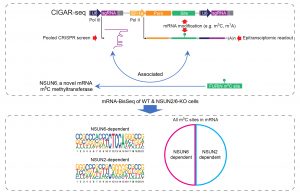
Fang, L.*#, W. Wang#, G. Li#, L. Zhang, J. Li, D. Gan, J. Yang, Y. Tang, Z. Ding, M. Zhang, W. Zhang, D. Deng, Z. Song, Q. Zhu, H. Cui, Y. Hu and W. Chen* (2020). “CIGAR-seq, a CRISPR/Cas-based method for unbiased screening of novel mRNA modification regulators.” Mol Syst Biol 16(11): e10025.
Yi, H.#, G. Li#, Y. Long, W. Liang, H. Cui, B. Zhang, Y. Tan, Y. Li, L. Shen, D. Deng, Y. Tang, C. Mao, S. Tian, Y. Cai, Q. Zhu, Y. Hu*, W. Chen* and L. Fang* (2020). “Integrative multi-omics analysis of a colon cancer cell line with heterogeneous Wnt activity revealed RUNX2 as an epigenetic regulator of EMT.” Oncogene 39(28): 5152-5164.
Praktiknjo, S. D.*#, B. Obermayer#, Q. Zhu#, L. Fang, H. Liu, H. Quinn, M. Stoeckius, C. Kocks, W. Birchmeier* and N. Rajewsky* (2020). “Tracing tumorigenesis in a solid tumor model at single-cell resolution.” Nat Commun 11(1): 991.
Li, Y.#, B. Schaefke*#, X. Zou, M. Zhang, F. Heyd, W. Sun, B. Zhang, G. Li, W. Liang, Y. He, J. Zhou, Y. Li, L. Fang, Y. Hu and W. Chen* (2020). “Pan-tissue analysis of allelic alternative polyadenylation suggests widespread functional regulation.” Mol Syst Biol 16(4): e9367.
Su, X., Y. Feng, S. A. Rahman, S. Wu, G. Li, F. Ruschendorf, L. Zhao, H. Cui, J. Liang, L. Fang, H. Hu, S. Froehler, Y. Yu, G. Patone, O. Hummel, Q. Chen, K. Raile, F. C. Luft, S. Bahring, K. Hussain, W. Chen, J. Zhang and M. Gong (2020). “Phosphatidylinositol 4-kinase beta mutations cause nonsyndromic sensorineural deafness and inner ear malformation.” J Genet Genomics 47(10): 618-626.
2019
Zhu, Q.#, L. Fang#, J. Heuberger, A. Kranz, J. Schipper, K. Scheckenbach, R. O. Vidal, D. Y. Sunaga-Franze, M. Muller, A. Wulf-Goldenberg, S. Sauer and W. Birchmeier* (2019). “The Wnt-Driven Mll1 Epigenome Regulates Salivary Gland and Head and Neck Cancer.” Cell Rep 26(2): 415-428 e415.
Wang, X., X. You, J. D. Langer, J. Hou, F. Rupprecht, I. Vlatkovic, C. Quedenau, G. Tushev, I. Epstein, B. Schaefke, W. Sun, L. Fang, G. Li, Y. Hu, E. M. Schuman and W. Chen* (2019). “Full-length transcriptome reconstruction reveals a large diversity of RNA and protein isoforms in rat hippocampus.” Nat Commun 10(1): 5009.
Qi, X. T., Y. L. Li, Y. Q. Zhang, T. Xu, B. Lu, L. Fang, J. Q. Gao, L. S. Yu, D. F. Zhu, B. Yang, Q. J. He and M. D. Ying (2019). “KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells.” Acta Pharmacol Sin 40(4): 546-555.
Lam, J. H., Y. Li, L. Zhu, R. Umarov, H. Jiang, A. Heliou, F. K. Sheong, T. Liu, Y. Long, Y. Li, L. Fang, R. B. Altman, W. Chen*, X. Huang* and X. Gao* (2019). “A deep learning framework to predict binding preference of RNA constituents on protein surface.” Nat Commun 10(1): 4941.
Chen, T., B. Zhang, T. Ziegenhals, A. B. Prusty, S. Frohler, C. Grimm, Y. Hu, B. Schaefke, L. Fang, M. Zhang, N. Kraemer, A. M. Kaindl, U. Fischer* and W. Chen* (2019). “A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing.” PLoS Genet 15(10): e1008460.
2018-earlier
Weng, Q., J. Wang, J. Wang, J. Wang, F. Sattar, Z. Zhang, J. Zheng, Z. Xu, M. Zhao, X. Liu, L. Yang, G. Hao, L. Fang, Q. R. Lu, B. Yang and Q. He (2018). “Lenalidomide regulates CNS autoimmunity by promoting M2 macrophages polarization.” Cell Death Dis 9(2): 251.
Schaefke, B.#, W. Sun#, Y. S. Li#, L. Fang and W. Chen (2018). “The evolution of posttranscriptional regulation.” Wiley Interdiscip Rev RNA 9(5): e1485.
Fang, L., Q. Zhu, M. Neuenschwander, E. Specker, A. Wulf-Goldenberg, W. I. Weis, J. P. von Kries and W. Birchmeier (2016). “A Small-Molecule Antagonist of the beta-Catenin/TCF4 Interaction Blocks the Self-Renewal of Cancer Stem Cells and Suppresses Tumorigenesis.” Cancer Res 76(4): 891-901.
Wend, P., L. Fang, Q. Zhu, J. H. Schipper, C. Loddenkemper, F. Kosel, V. Brinkmann, K. Eckert, S. Hindersin, J. D. Holland, S. Lehr, M. Kahn, U. Ziebold and W. Birchmeier (2013). “Wnt/beta-catenin signalling induces MLL to create epigenetic changes in salivary gland tumours.” EMBO J 32(14): 1977-1989.
Holland, J. D., B. Gyorffy, R. Vogel, K. Eckert, G. Valenti, L. Fang, P. Lohneis, S. Elezkurtaj, U. Ziebold and W. Birchmeier (2013). “Combined Wnt/beta-catenin, Met, and CXCL12/CXCR4 signals characterize basal breast cancer and predict disease outcome.” Cell Rep 5(5): 1214-1227.
Jia, P., R. Sheng, J. Zhang, L. Fang, Q. He, B. Yang and Y. Hu (2009). “Design, synthesis and evaluation of galanthamine derivatives as acetylcholinesterase inhibitors.” Eur J Med Chem 44(2): 772-784.
Xu, D., L. Fang, Q. Zhu, Y. Hu, Q. He and B. Yang (2008). “Antimultidrug-resistant effect and mechanism of a novel CA-4 analogue MZ3 on leukemia cells.” Pharmazie 63(7): 528-533.
Weng, Q., D. Wang, P. Guo, L. Fang, Y. Hu, Q. He and B. Yang (2008). “Q39, a novel synthetic Quinoxaline 1,4-Di-N-oxide compound with anti-cancer activity in hypoxia.” Eur J Pharmacol 581(3): 262-269.
Fang, L., L. Shen, Y. Fang, Y. Hu, Q. He and B. Yang (2008). “MZ3 can induce G2/M-phase arrest and apoptosis in human leukemia cells.” J Cancer Res Clin Oncol 134(12): 1337-1345.
Fang, L., Q. He, Y. Hu and B. Yang (2007). “MZ3 induces apoptosis in human leukemia cells.” Cancer Chemother Pharmacol 59(3): 397-405.
Yang, B., L. Fan, L. Fang and Q. He (2006). “Hypoxia-mediated fenretinide (4-HPR) resistance in childhood acute lymphoblastic leukemia cells.” Cancer Chemother Pharmacol 58(4): 540-546.
He, Q., R. Li, L. Fang, H. Ying, Y. Hu and B. Yang (2006). “Antileukemia activity of MSFTZ-a novel flavanone analog.” Anticancer Drugs 17(6): 641-647.
加入团队
联系方法
邮箱:fangl@sustech.edu.cn
电话:13823288350